Section: New Results
Medical Image Analysis
Segmentation and Anatomical Variability of the Cochlea from Medical Images
Participants : Thomas Demarcy [Correspondant] , Hervé Delingette, Clair Vandersteen [IUFC, Nice] , Dan Gnansia [Oticon Medical] , Nicholas Ayache.
This work is supported by the National Association for Research in Technology (ANRT) through the CIFRE Grant 2013-1165 and Oticon Medical (Vallauris). Part of this work is also funded by the European Research Council through the ERC Advanced Grant MedYMA 2011-291080 (on Biophysical Modeling and Analysis of Dynamic Medical Images). This work is a collaboration with the Department of Ear Nose Throat Surgery (IUFC, Nice) and the Nice University Hospital (CHU).
Image segmentation, Surgery planning, Shape modeling, Anatomical variability, Cochlear implant, Temporal bone.
-
We evaluated the optimal electrode diameter in relation to the cochlear shape [20].
-
We proposed a novel framework for estimating the insertion depth and its uncertainty from segmented CT images based on a new parametric shape model [37].
-
We provided a proof of concept for the estimation of postoperative cochlear implant electrode-array position from clinical CT [44] (Fig. 1).
|
Infarct Localization and Uncertainty Quantification from Myocardial Deformation
Participants : Nicolas Duchateau [Correspondant] , Maxime Sermesant.
This work received the partial support from the European Union 7th Framework Programme (VP2HF FP7-2013-611823) and the European Research Council (MedYMA ERC-AdG-2011-291080).
Myocardial infarct, Computer-aided diagnosis, Dimensionality reduction, Biomechanical modeling.
-
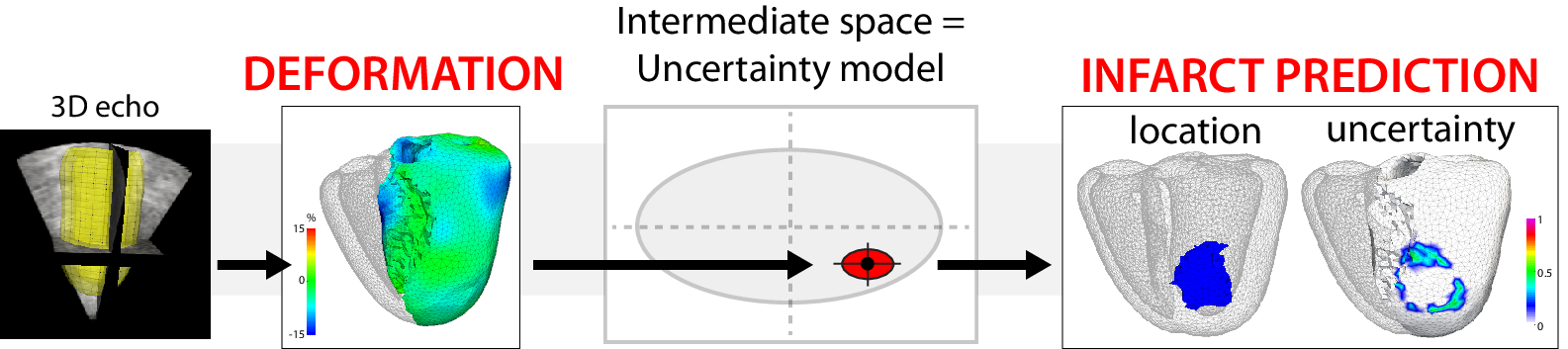
We build upon preliminary work for the automatic localization of myocardial infarct from local wall deformation, which has potential for risk stratification from routine examination such as 3D echocardiography. Non-linear dimensionality reduction serves to estimate the Euclidean space of coordinates encoding deformation patterns (training phase), and is combined with multi-scale kernel regressions to link the deformation patterns, the low-dimensional coordinates and the infarct location for new cases (testing phase).
-
We extend this approach by considering the different components of myocardial strain considered in clinical practice, and by taking advantage of the space of low-dimensional coordinates to model uncertainty in the infarct localization [18].
-
These concepts were tested on 500 synthetic cases with infarcts of random extent, shape, and location, generated from a realistic electromechanical model, and 108 pairs of 3D echocardiographic sequences and delayed-enhancement magnetic resonance images from real cases. Infarct prediction is made at a spatial resolution more than 10 times smaller than the current diagnosis, made regionally. Our method is accurate, and significantly outperforms the clinically-used thresholding of the deformation patterns. Uncertainty adds value to refine the diagnosis and eventually re-examine suspicious cases.
|
Longitudinal Analysis and Modeling of Brain Development
Participants : Mehdi Hadj-Hamou [Correspondant] , Xavier Pennec, Nicholas Ayache, Hervé Lemaitre [Inserm U1000] , Jean-Luc Martinot [Inserm U1000] .
This work is partly funded through the ERC Advanced Grant MedYMA 2011-291080 (on Biophysical Modeling and Analysis of Dynamic Medical Images).
Processing pipeline, Brain development, Adolescence, Longitudinal analysis, Non-rigid registration algorithm, Extrapolation.
-
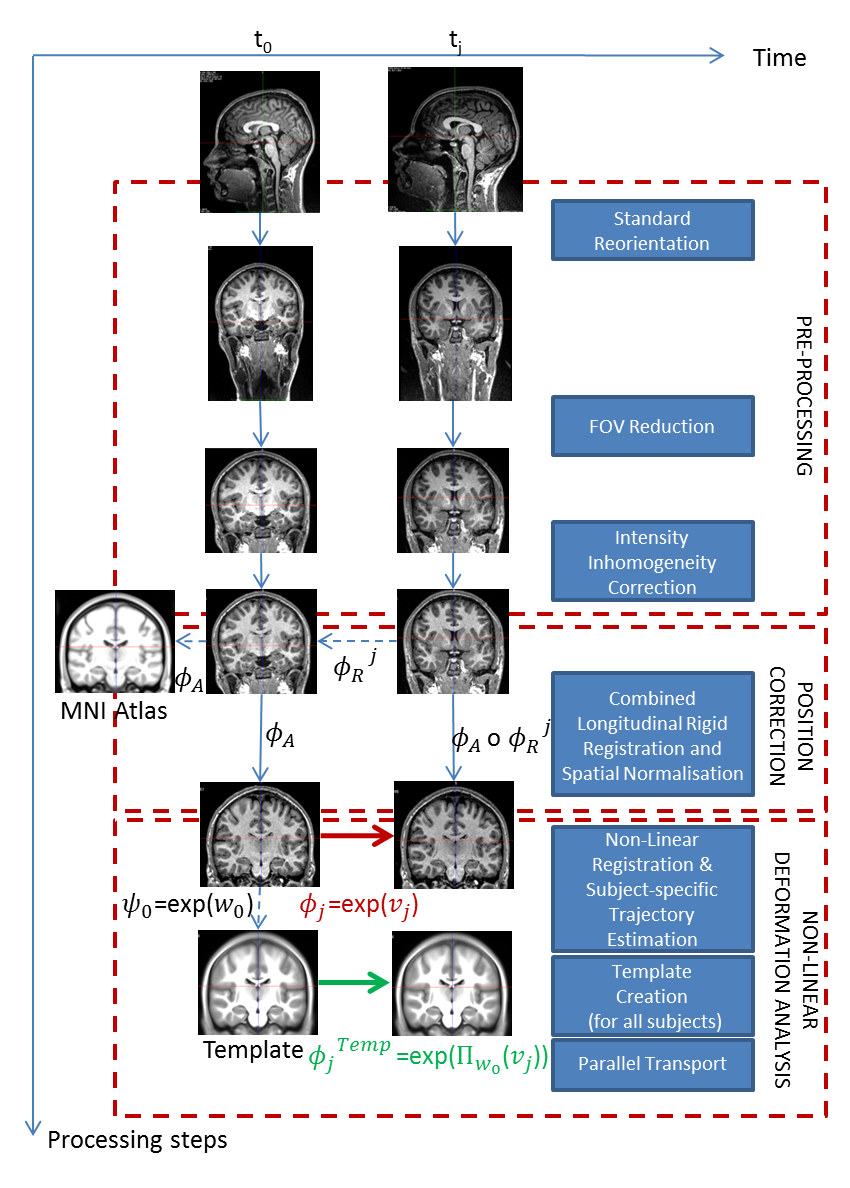
We propose and detail a deformation-based morphometry computational framework, called Longitudinal Log-Demons Framework (LLDF), to estimate the longitudinal brain deformations from image data series, transport them in a common space and perform statistical group-wise analyses. It is based on freely available software and tools, and consists of three main steps (cf. Fig. 3):
It is based on the LCC log-Demons non-linear symmetric diffeomorphic registration algorithm with an additional modulation of the similarity term using a confidence mask to increase the robustness with respect to brain boundary intensity artifacts.
-
This work led to a published journal publication [23].
-
The LLDF pipeline is exemplified on the longitudinal Open Access Series of Imaging Studies (OASIS) database and is applied to the study of longitudinal trajectories during adolescence, for which little is known. The aim of this project is to provide models of brain development during adolescence based on diffeomorphic registration parametrised by SVFs. We particularly focused our study on the link between sexual dimorphism and the longitudinal evolution of the brain. This work was done in collaboration with J.L. Martinot et H. Lemaître (Inserm U1000).
Left Atrial Wall Segmentation and Thickness Measurement using Region Growing and Marker-Controlled Geodesic Active Contour
Participants : Shuman Jia [Correspondant] , Loïc Cadour, Hubert Cochet [IHU Liryc, Bordeaux] , Maxime Sermesant.
The authors acknowledge the partial funding by the Agence Nationale de la Recherche (ANR)/ERA CoSysMed SysAFib and ANR MIGAT projects.
Atrial fibrillation, Left atrial wall thickness, Image segmentation, Cardiac computed tomography (CT), Region growing, Geodesic active contour.
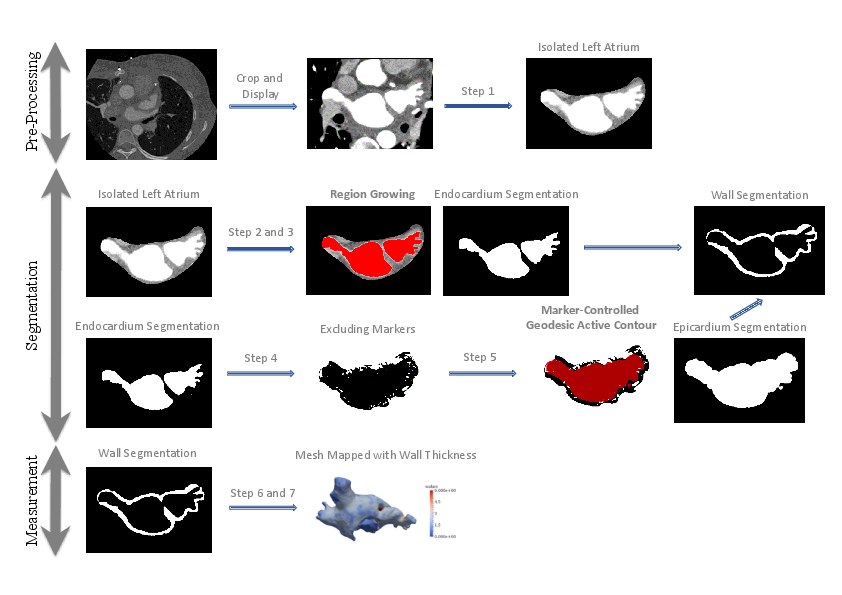
We proposed a method to segment the left atrial (LA) wall and measure the wall thickness from cardiac computed tomography images, making use of patient-specific intensity value information and surrounding environment (see Fig. 4).
We partially implemented the method in the MUSIC software and tested our pipeline on 10 datasets. The results achieved a good match of wall thickness with manual segmentation. We received a Best Paper Award for this work [39] at the 2016 STACOM Workshop in Athens, Greece.
Weakly Supervised Learning for Tumor Segmentation
Participants : Pawel Mlynarski [Correspondant] , Nicholas Ayache, Hervé Delingette, Antonio Criminisi [MSR] .
This work is funded by the Inria-Microsoft joint center and is done in cooperation with Microsoft Research in Cambridge.
Deep Learning, Segmentation, Classification, Tumor.
-
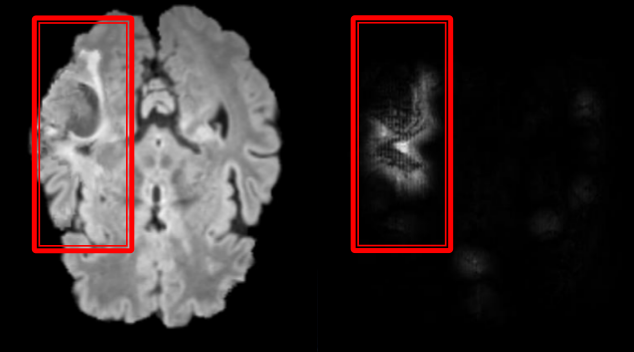
The goal of this work is to develop new machine learning methods for the localization and segmentation of tumors, without relying on the ground truth provided by experts. In particular, we study Deep Learning methods for classification and weakly supervised localization (Figure 5).
-
We proposed two methods of synthesis of brain 3D MR images, in order to use them during the training of Neural Nets. The proposed methods showed to improve our performance in localization of brain tumors.
|
Inter-Operative Relocalization in Flexible Endoscopy
Participants : Anant Vemuri [Correspondant] , Stéphane Nicolau, Luc Soler, Nicholas Ayache.
This work has been performed in collaboration with IHU Strasbourg and IRCAD, France.
Computer-assisted intervention, Barrett's esophagus, Biopsy relocalization, Electromagnetic tracking.
Oesophageal adenocarcinoma arises from Barrett’s oesophagus, which is the most serious complication of gastro-oesophageal reflux disease. Strategies for screening involve periodic surveillance and tissue biopsies. A major challenge in such regular examinations is to record and track the disease evolution and relocalization of biopsied sites to provide targeted treatments.
In support of this work, the thesis [6] was defended before a committee of medical experts and scientific reviewers, on April 26th 2016.
References:
-
Vemuri, Nicolau, Sportes, Marescaux, Soler, Ayache. Inter-Operative Biopsy Site Relocalization in Endoluminal Surgery [35].
-
Nicolau, Vemuri, Soler, Marescaux. Anatomical site relocalisation using dual data synchronisation (patent) [49].
Learning Brain Alterations in Multiple Sclerosis from Multimodal Neuroimaging Data
Participants : Wen Wei, Nicholas Ayache, Stanley Durrleman [ARAMIS] , Olivier Colliot [ARAMIS] .
Multiple sclerosis, Neuroimageing.
The goal of this topic is to develop a machine learning approach that can predict different types of PET-derived brain alterations using multiple local and regional MRI measures.
Deep Learning for Cardiac Image Analysis
Participants : Qiao Zheng [Correspondant] , Hervé Delingette, Nicholas Ayache.
Deep learning, Artificial neural network, Cardiac image.
Deep learning has proven to be very successful in computer vision and image understanding. However, its potential for medical image analysis has yet to be explored. We apply deep learning on cardiac images in order to learn cardiac image processing and anomaly detection. Our work includes data collection and preprocessing, software engineering, learning process design, etc.